SMS 2023 Spring Image Competition
Images of Research
Well done to Dr Mattias Malaguti, a Senior Researcher at the University of Edinburgh for their winning entry of ‘Hair Follicle Rainbow’. Mattias won the £100 prize
Winner: Hair follicle rainbow
Dr Mattias Malaguti, University of Edinburgh
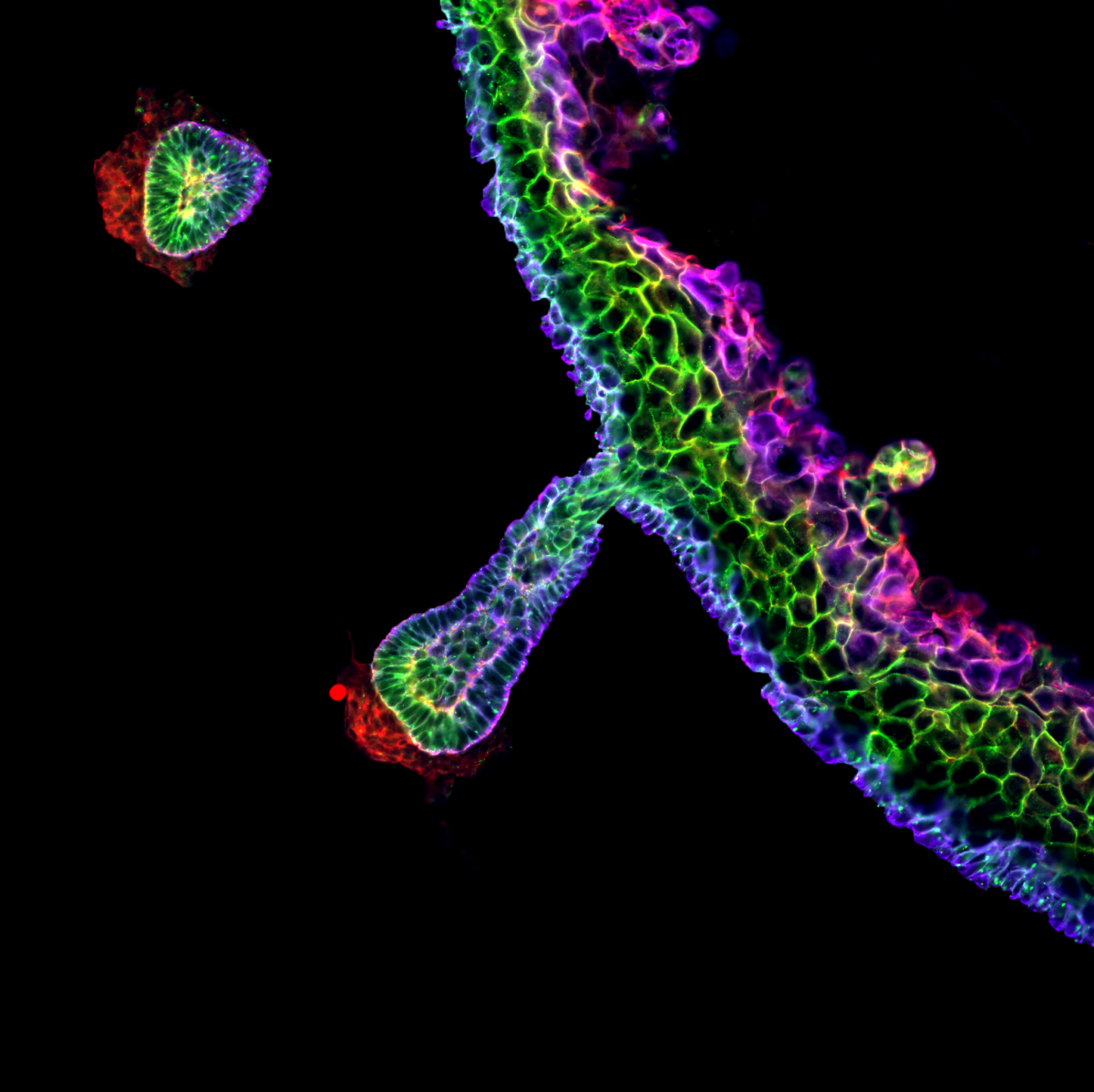
The image is of a skin organoid derived entirely from human pluripotent cells, fixed 84 days after the start of the differentiation protocol. This section was stained for the tight junction marker ZO-1 (red), the adherens junction protein E-cadherin (green), and the basal keratinocyte marker Keratin 14 (blue). The different patterns of expression of these proteins create a “rainbow” effect in the stratified epidermis, as well as in the hair follicle, which produces pigmented hairs. The red staining underlying the base of the hair follicle marks the location of the dermal papilla. This image was acquired on a Nikon TiE Eclipse widefield microscope in the Institute for Regeneration and Repair Imaging Facility, with a 20X apochromatic lens. Background staining in the dermis was selectively removed in Photoshop for artistic effect. The image was obtained as a collaboration between myself, Eleanor Earp and Maria Rosa Portero Migueles.
Competition Entries
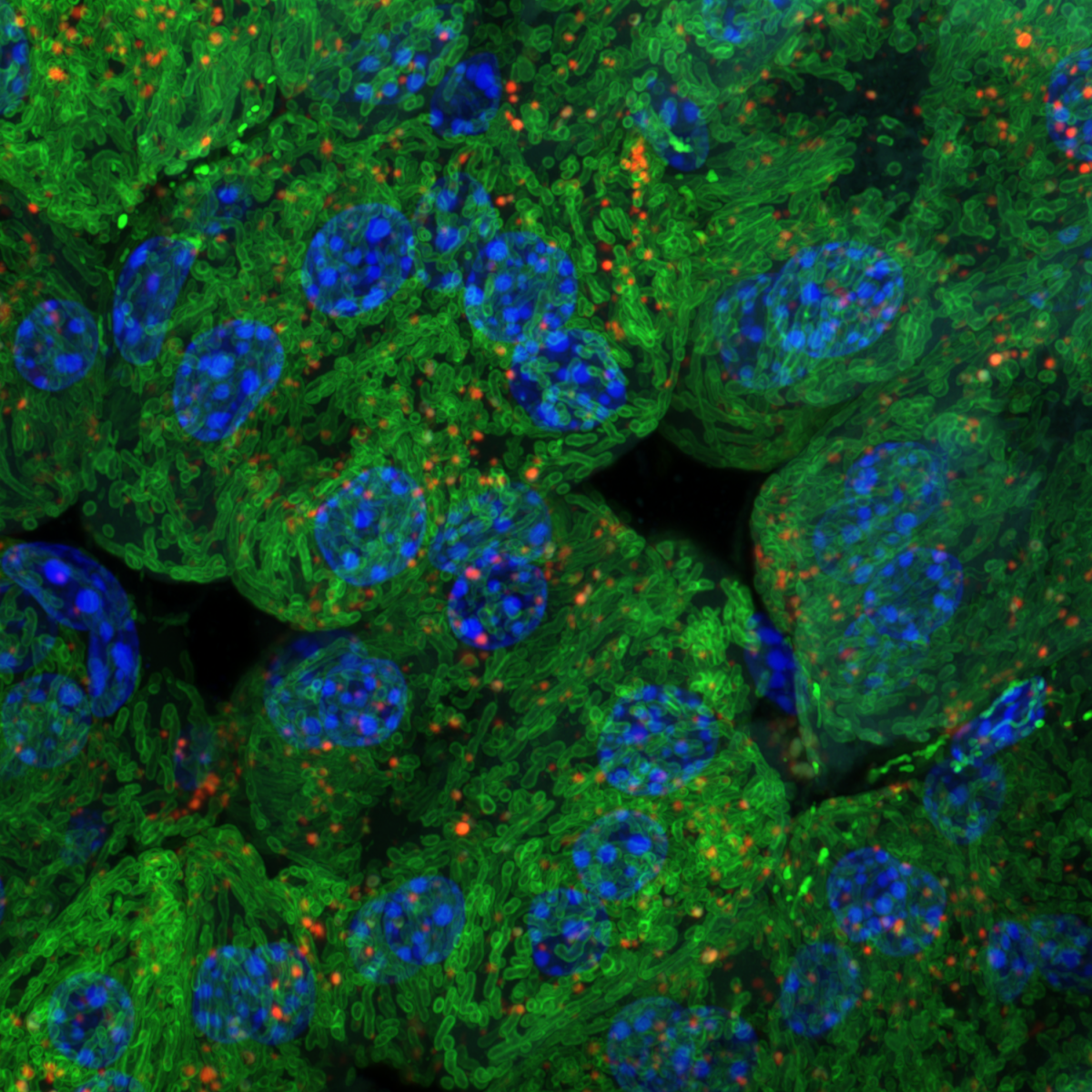
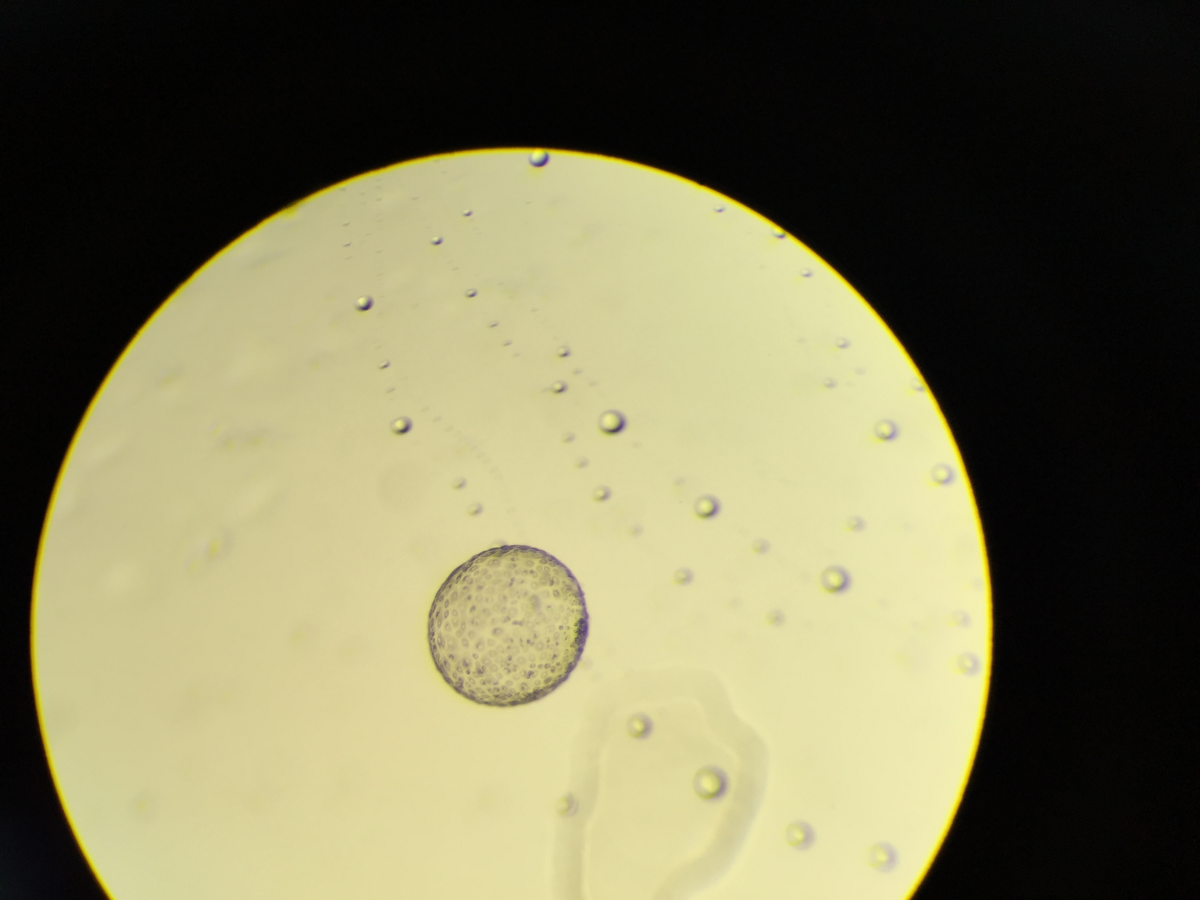
1. Power Lunch
Dr François Singh, University of Dundee
Pancreatic acini from mice expressing the mito-QC reporter, enabling to visualise mitophagy in vivo. Green: Mitochondria Red: Mitolysosome Blue: Nuclei (Hoechst 33342) Super-resolution maximal intensity projection of 3D z-stacks obtained with a Zeiss 880 LSM with Airyscan (63x objective). Imaged at the Dundee Imaging Facility (DIF)
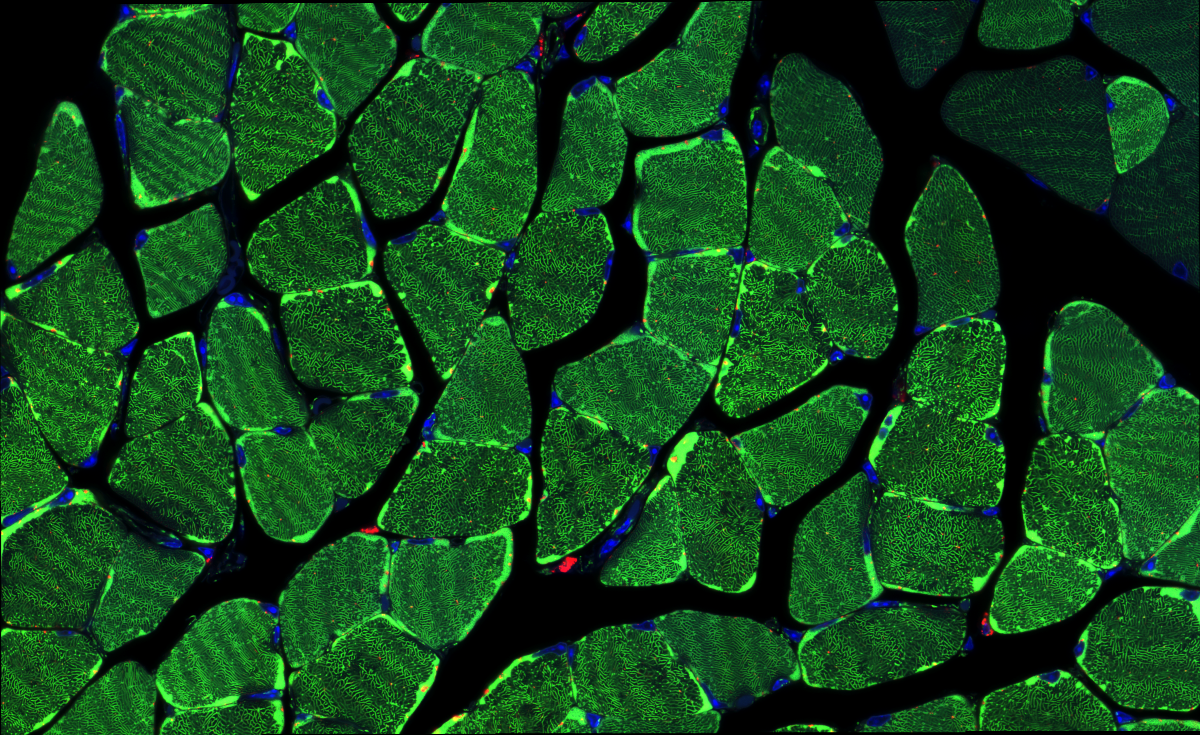
2. Soleal Labyrinth
Dr François Singh, University of Dundee
Soleus muscle cross-section from mice expressing the mito-QC reporter, enabling to visualise mitophagy in vivo. Green: Mitochondria Red: Mitolysosomes Blue: Nuclei (Hoechst 33342) Super-resolution tile scan obtained with a Zeiss 880 LSM with Airyscan (63x objective). Imaged at the Dundee Imaging Facility (DIF)
3. Parasite Petals
Dr Emma Sands, University of Dundee
Cryptosporidium is an intestinal diarrhoea-causing parasite. Cryptosporidiosis is a leading cause of infant diarrhoea, but there are no approved vaccines or drugs. These parasites lurk in contaminated water, contained within an egg shell. Malnourished children get especially sick, and communities who lack access to clean drinking water, or share drinking water with livestock being particularly at risk.
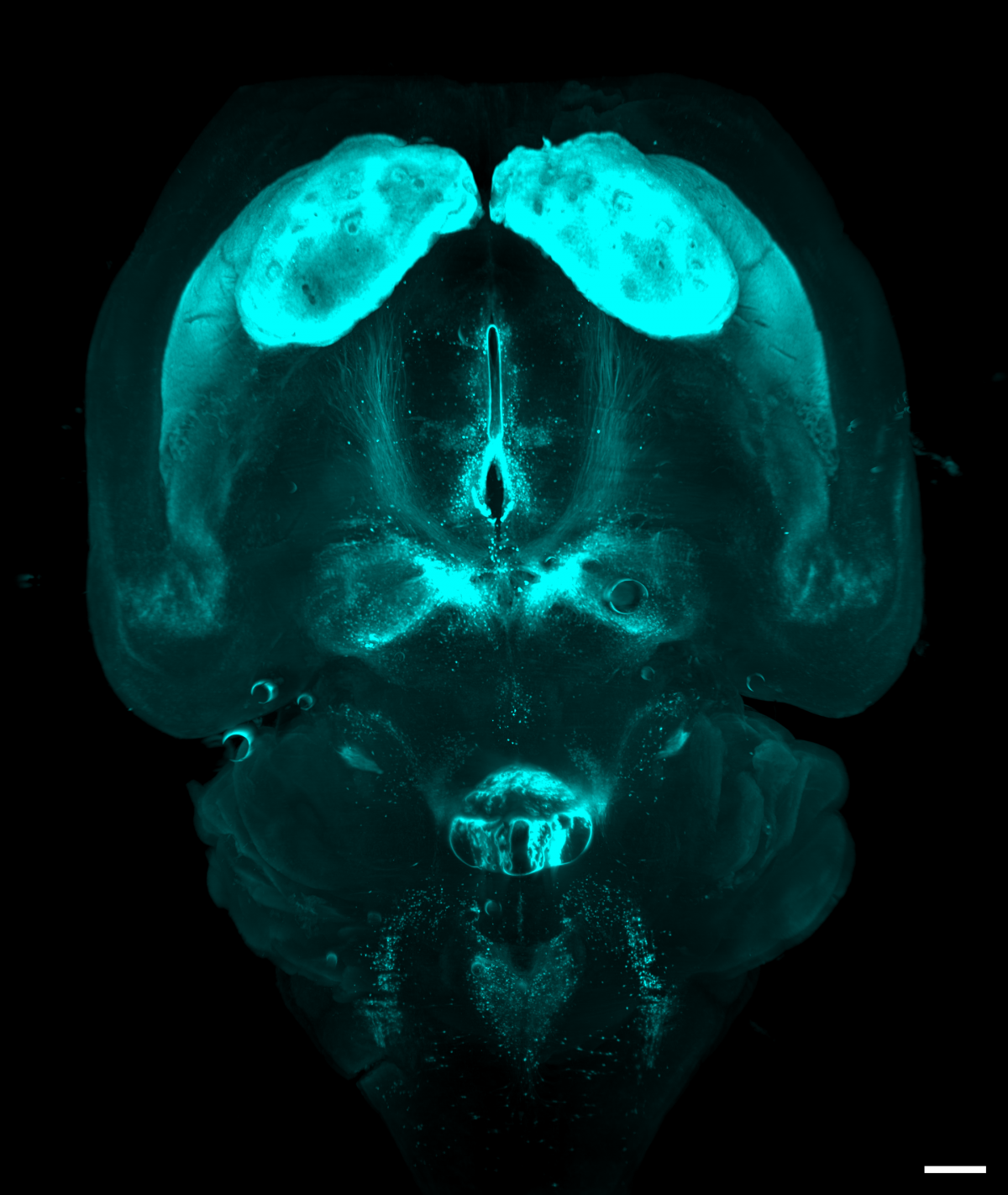
4. Mobilize and love your brain
Dr Cristina Martinez Gonzalez, University of Edinburgh
Light sheet microscopy images of a whole mouse brain that was optically cleared using a RatDISCO protocol and immunolabelled against Tyrosine hydroxylase. Images were acquired in the horizontal plane using an Ultramicroscope II (maximum intensity projection; single field of view; imaging time 30 min) with an Olympus MVPLAPO 2x objective, working distance 6 mm; Andor Neo sCMOS camera and a 561nm laser. The image was adjusted in FIJI for brightness and contrast and assigned cyan LUT; scale bar 500 um.
Tyrosine hydroxylase (TH) labels dopaminergic, adrenergic and norepinephrinergic neurons in the brain and medulla oblongata involved in reward, motor control and mobilizing the brain and body for action.
In the image, dopaminergic neurons in the substantia nigra pars compacta and the ventral tegmental area send ascending projections that innervate a large number of subcortical, cortical structures and the basal ganglia, in particular the striatum, which receives the densest dopaminergic input. Adrenergic and norepinephrinergic neurons in the brainstem formed a heart-shaped silhouette at the bottom of the image.
Imaging the whole brain without mechanical sectioning allowed us to rapidly image, identify and quantify TH-immunopositive neurons and their projections in several experimental animals. Light sheet microscopy provides comprehensive, novel, unbiased (and beautiful) information that traditional methods cannot achieve.
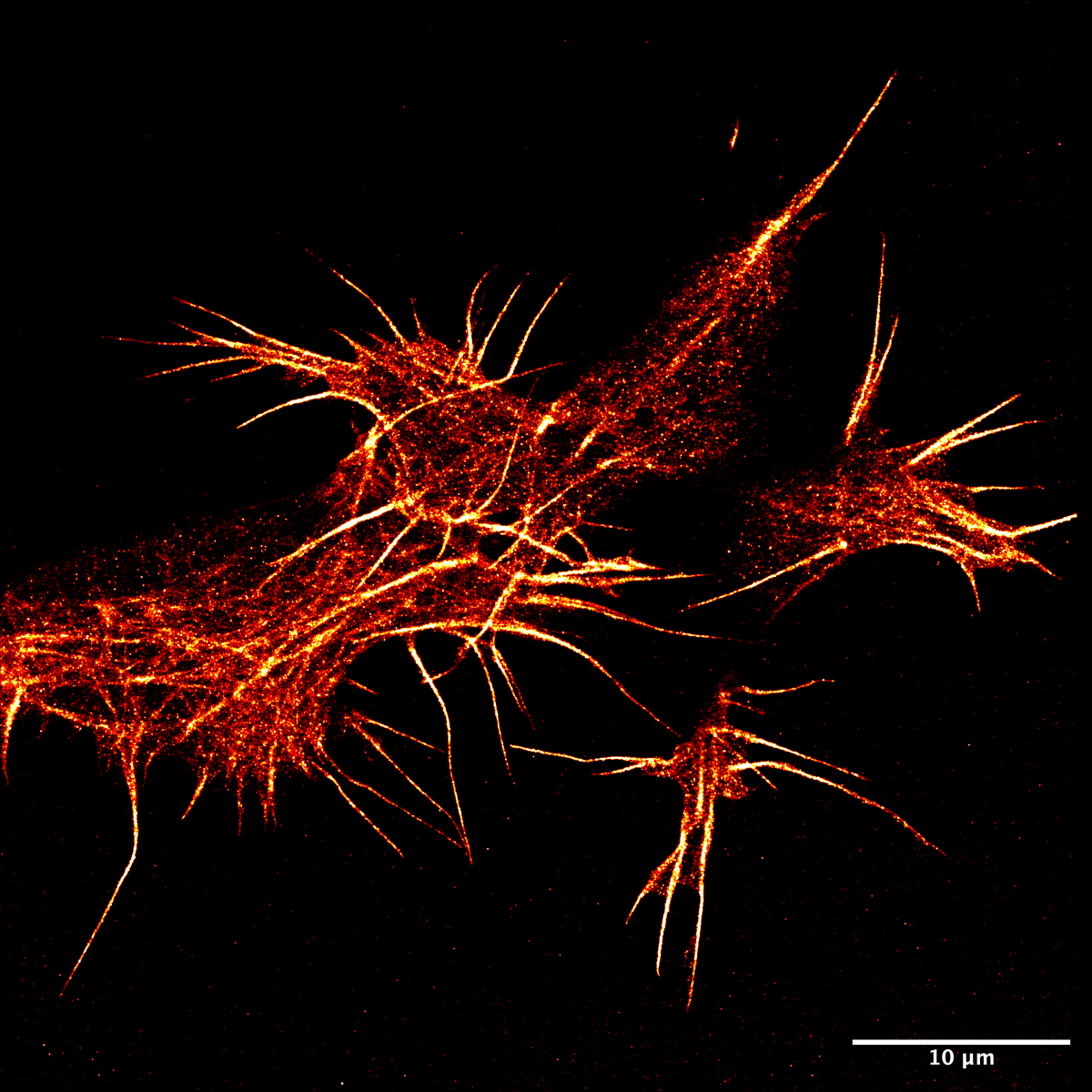
5. F-actin in super-resolution in neuroblastoma
Haresh Bhaskar, University of Edinburgh
Actin is an abundant protein found in all animal cells and plays a vital role in maintaining cell shape. This filamentous protein has always proven difficult to image in live cells due to fluorescent protein fusions causing disruptions to their polymerisation dynamics. Using a novel PAINT-based technique in a TIRF system and the LifeAct-14 probe, we have imaged actin in super-res in live SH-SY5Y cells.
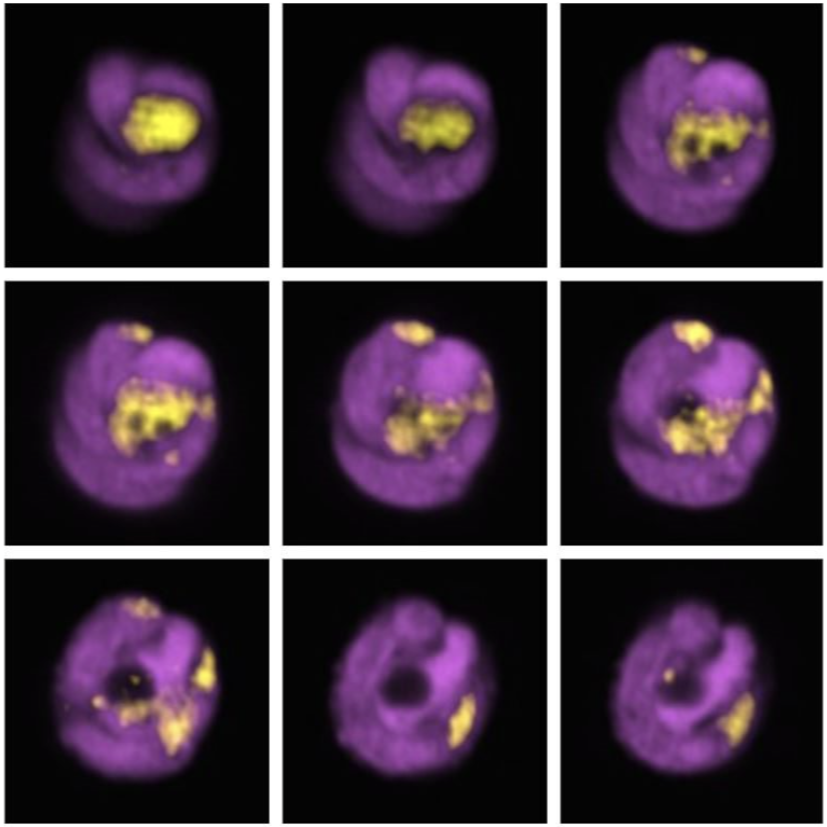
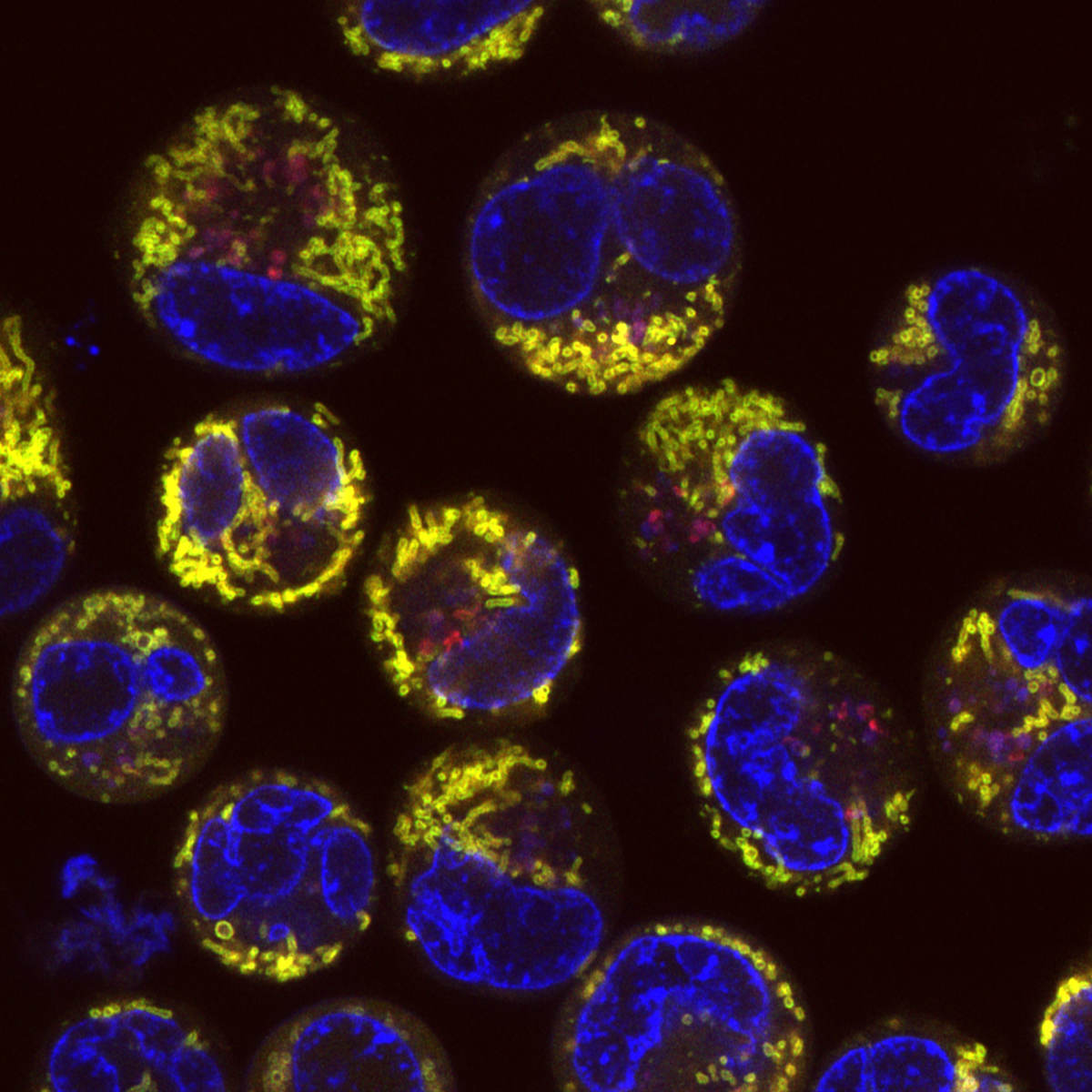
6. Mitophagy in Leukaemia Cells
Desiree Zerbst, University of Glasgow
Live cell imaging of basal mitophagy in human leukaemia cells expressing the oncogene BCR-ABL using the MitoQC reporter gene, which encodes for the tandem dye GFP-mCherry that localises to the outer mitochondrial membrane. Healthy mitochondria appear in yellow due to the GFP and mCherry overlapping. Mitochondria that are degraded via the autophagosomal pathway lose their GFP signal as it is quenched due to the low pH in the lysosome. This allows to observe healthy and damaged mitochondria undergoing mitophagy. Live cells are imaged with the Zeiss LSM 880 with confocal super-resolution (Airyscan) using a 63x objective. The nuclei is shown in blue (DAPI), healthy mitochondria give both GFP and mCherry signal resulting in a yellow colour and damaged mitochondria are detected in red (mCherry). Images were acquired in the BAIR facility of the Beatson Institute for Cancer Research in Glasgow.
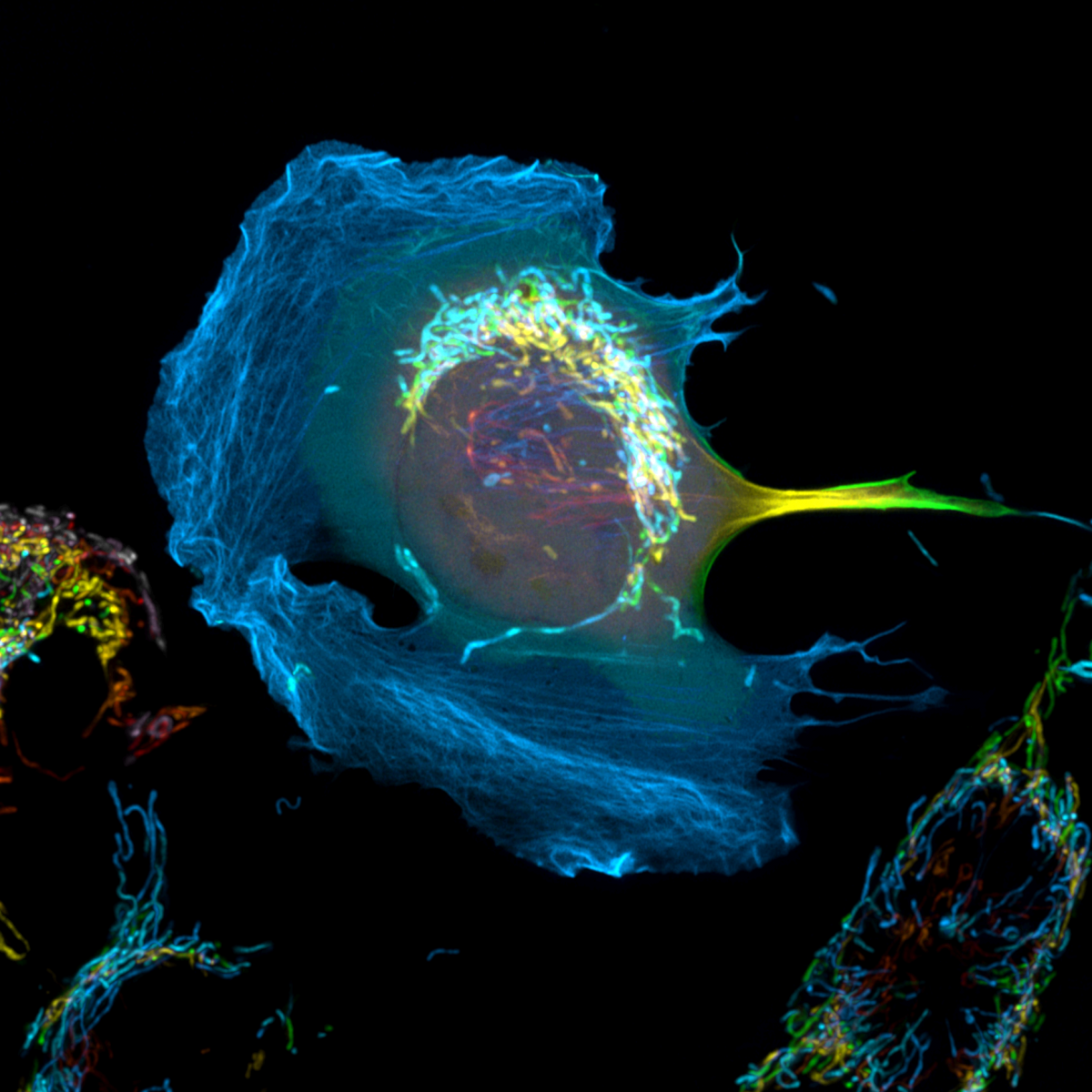
7. Lost in Space
Dr Peggy Paschke, CRUK Beatson Institute
Hcmel melanoma cells stably expressing cyto/lbnox were transfected with Lifeact-mClover2 to visualize their actin cytoskeleton. The expression of cyto/lbnox changes the redox status of the cells but also results in the formation of large lamellipodia. Additionally, the mitochondria were labelled with PK-mito deep red. Shown is the maximum intensity projection of a Z-stack in which each plane is labelled in a different colour. Zeiss 880 Airyscan, 63x Oil objective, SR-mode.
8. Mitochondrial Forest
Dr Peggy Paschke, CRUK Beatson Institute
Hcmel melanoma cells stably expressing cyto/lbnox were transfected with Lifeact-mClover2 to visualize their actin cytoskeleton. The expression of cyto/lbnox changes the redox status of the cells but also results in the formation of large lamellipodia. Additionally, the mitochondria were labelled with PK-mito deep red. Shown is a maximum intensity protection of a timelapse movie that was acquired over 15 minutes – with one frame taken every 30 seconds. Zeiss 880 Airyscan, 63x Oil objective, SR-mode.
9. Hair follicle rainbow
Dr Mattias Malaguti, University of Edinburgh
The image is of a skin organoid derived entirely from human pluripotent cells, fixed 84 days after the start of the differentiation protocol. This section was stained for the tight junction marker ZO-1 (red), the adherens junction protein E-cadherin (green), and the basal keratinocyte marker Keratin 14 (blue). The different patterns of expression of these proteins create a “rainbow” effect in the stratified epidermis, as well as in the hair follicle, which produces pigmented hairs. The red staining underlying the base of the hair follicle marks the location of the dermal papilla. This image was acquired on a Nikon TiE Eclipse widefield microscope in the Institute for Regeneration and Repair Imaging Facility, with a 20X apochromatic lens. Background staining in the dermis was selectively removed in Photoshop for artistic effect. The image was obtained as a collaboration between myself, Eleanor Earp and Maria Rosa Portero Migueles.
10. Calcium activity during whisker stimulation
Dr Juraj Koudelka , University of Edinburgh
Timelapse of a whisker stimulation response in an awake mouse barrel cortex through a cranial window. Brain vasculature is visualised via intravenous injection of Rhodamine B dextran (red); astrocytic calcium signalling via intracortical injection of an astrocyte-specific GCaMP-AAV (green) at 25X using Leica SP8 DIVE 2-photon microscope. The mouse has been habituated to the imaging set up and is free-running on a treadmill, while its whiskers are mechanically stimulated at three different timepoints for 30 seconds at 6Hz each (on the video at 1:00, 2:30 and 4:00). The timelapse recording (at 512px x 512px, 4.3fps) can then be used to monitor changes in individual blood vessels and investigate changes in astrocytic calcium signalling via the GCaMP fluorescence intensity. Video has been smoothed in FIJI, otherwise not altered. Scale bar 50um
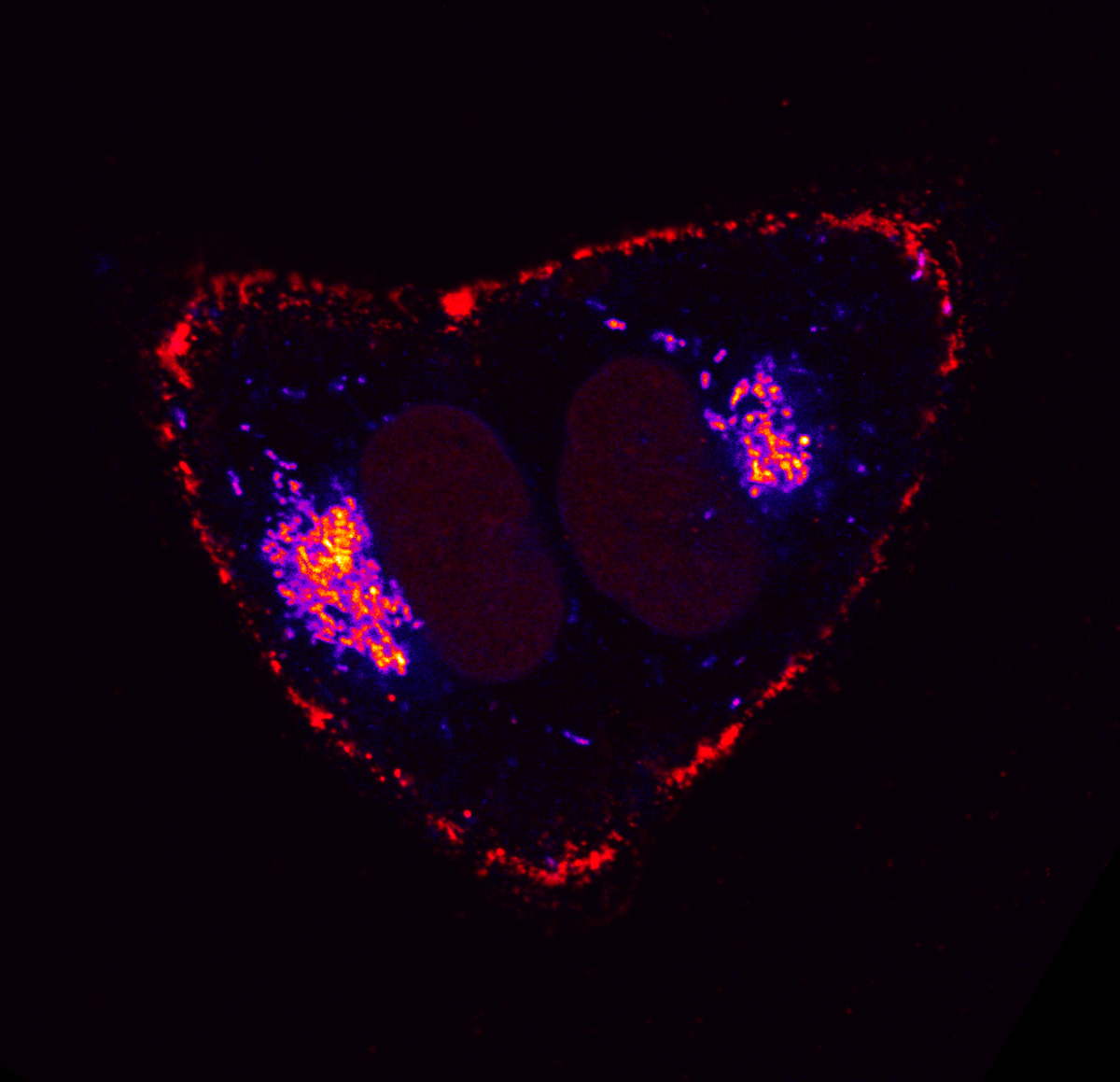
11. Sending Love
Dr Rosalie Heilig, Beatson Institute Glasgow
1. Two cells dying of apoptosis. The cells were treated to undergo apoptosis and stained for mitochondria as well as DNA. The cells were captured at the right moment before rounding up and permeabilising. 2. Confocal live cell imaging (timelapse), LSM880, 63 oil objective, 488, 647 3. Mouse endothelial cells 4. BAIR, Beatson Cancer Institute
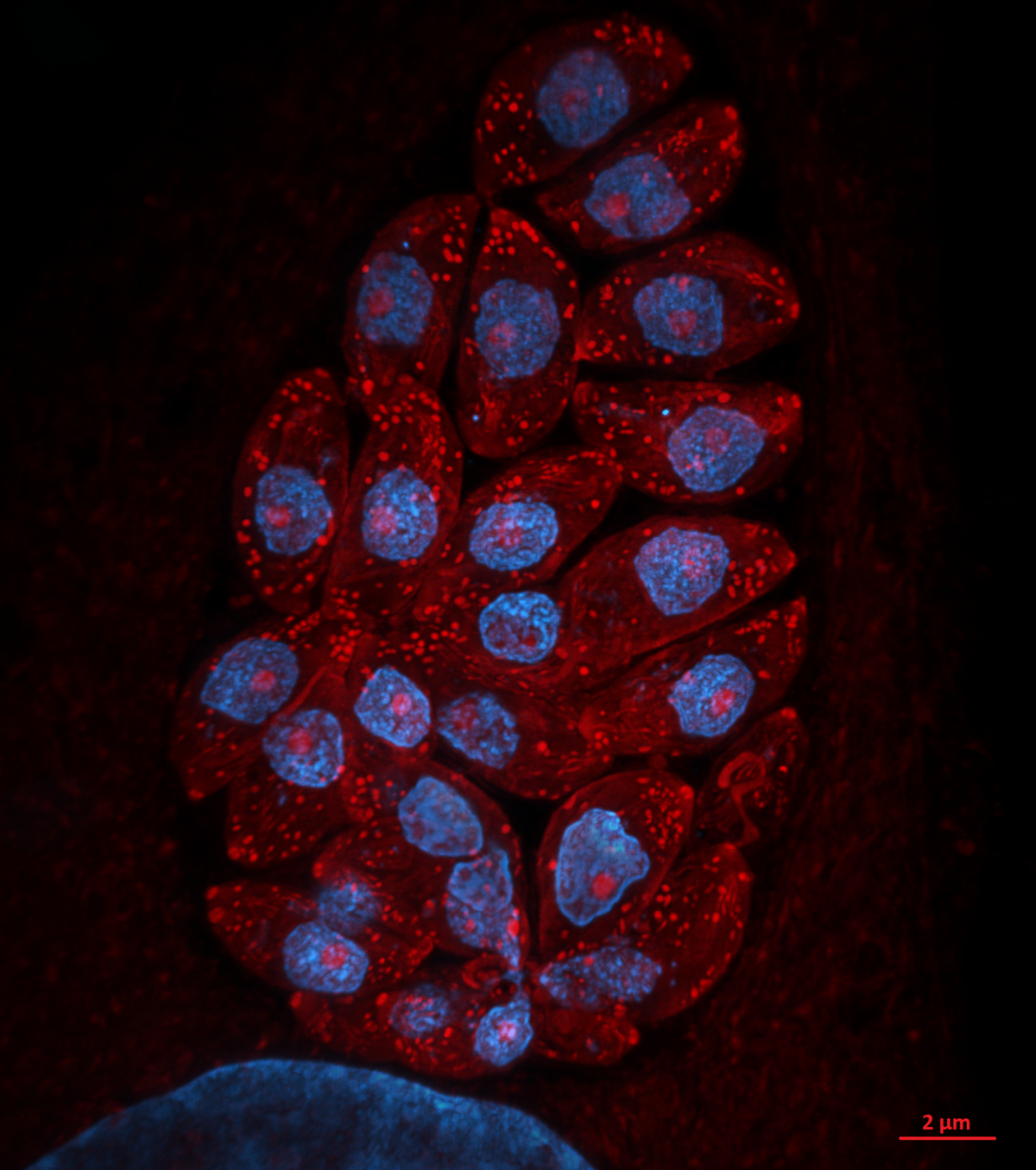
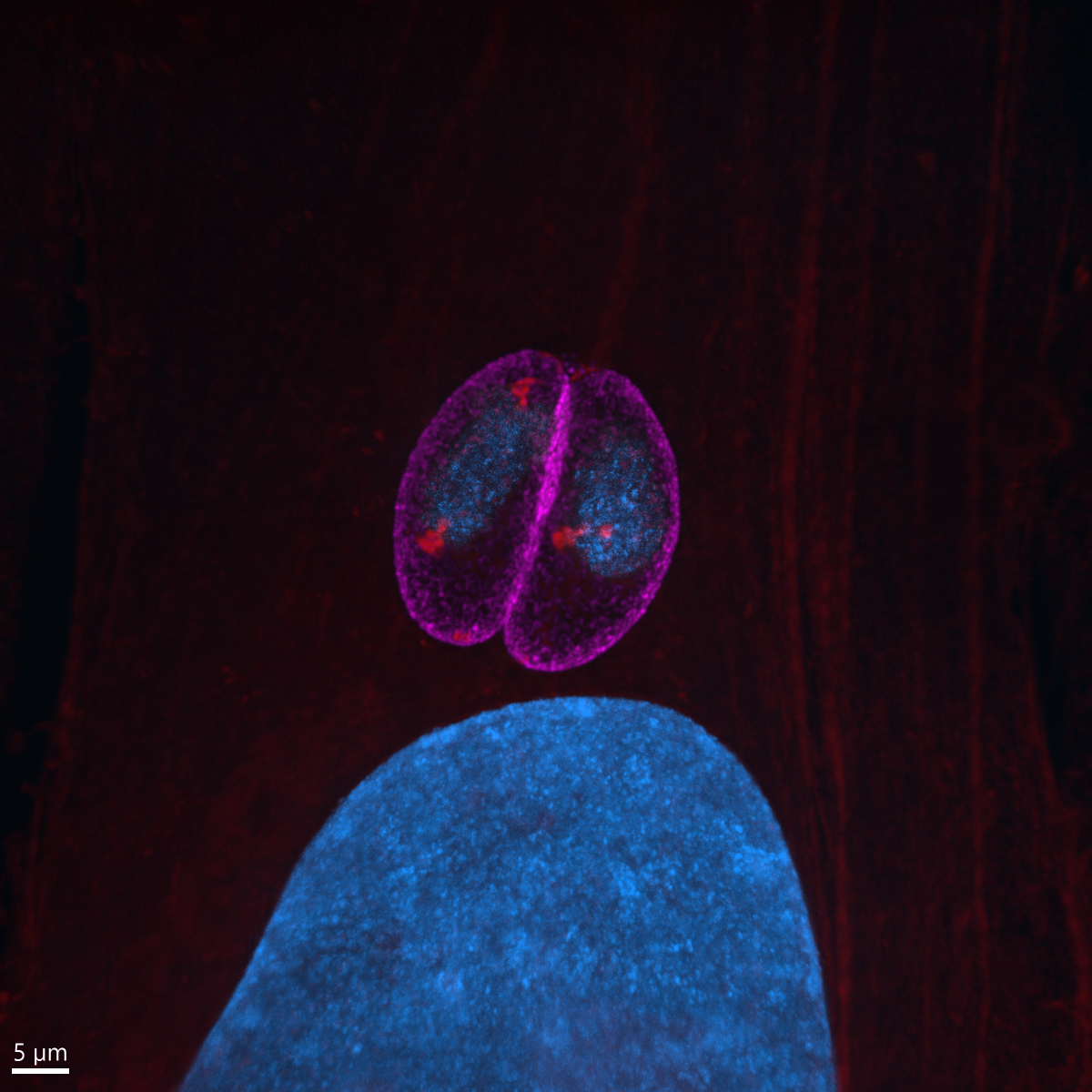
12. Toxoplasma pinecone
Dr Kseniia Bondarenko, University of Edinburgh
Admire the Toxoplasma gondii parasites growing inside a human skin cell next to its big blue nuclei! The protein-rich parasitic organelles are visible in bright red (594-NHS-ester) and nuclei in blue (DNA stain). Infected cells were gelled and expanded 4.5x using Ultrastructure Expansion Microscopy (U-ExM) and imaged with Nikon Ti2 CSU-W1 Spinning Disk confocal microscope, 100x, at the COIL imaging facility. Expansion Microscopy is a relatively new technique which allows increasing the spatial resolution within a cell or tissue for microscopic imaging through the physical expansion of the sample, bringing super-resolution quality to regular epifluorescence or confocal microscopes (instead of deploying time-consuming and expensive super-resolution techniques like STED and STORM). U-ExM allows detailed investigation of protein localization in Toxoplasma parasites with a spatial resolution of ~65 nm using a conventional confocal setup. We use it to clarify and discover new locations of proteins that are either upregulated or change their location when switching to the chronic state of toxoplasmosis. Model organism: human foreskin fibroblasts infected with ME49 strain of Toxoplasma gondii. Prep: infected cells were fixed 24h post-infection using 4% formaldehyde and then stained with non-selective protein marker N-hydroxysuccinimide (NHS)-ester conjugated to AlexaFluor 594, and Hoechst 33342 nucleic acid stain.
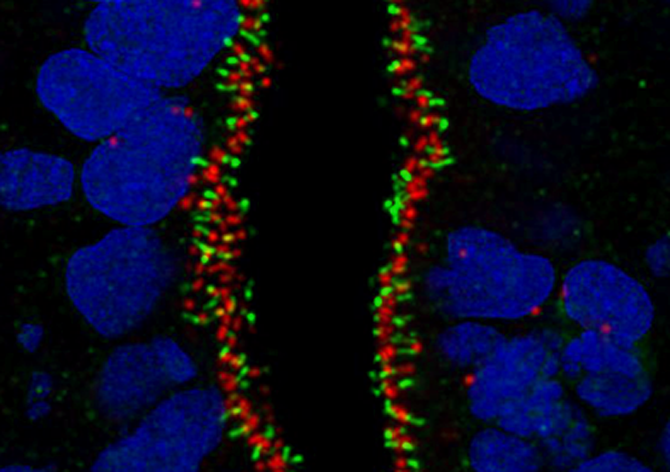
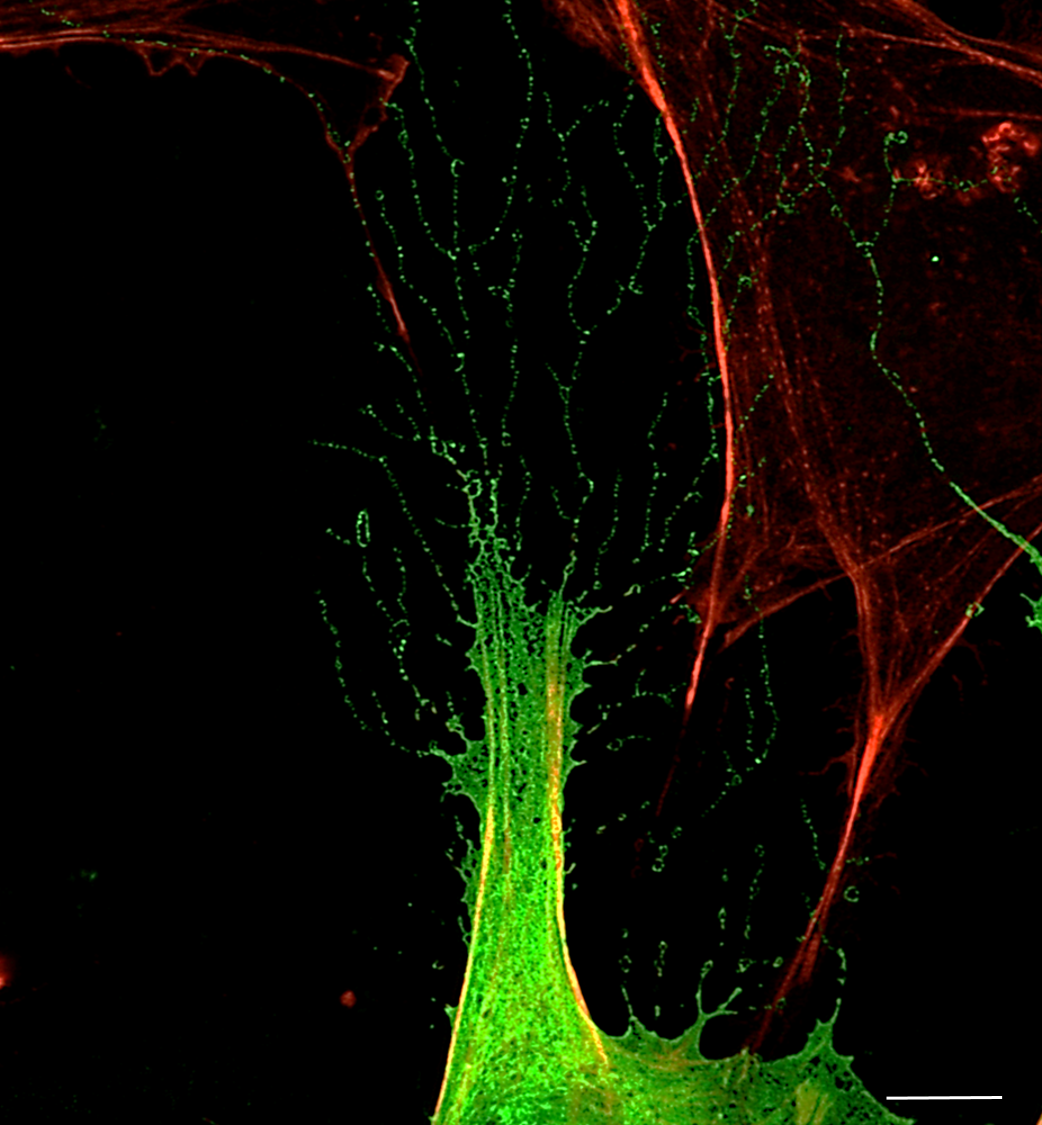
13. Human antennas - you can never have enough of them
Dr Thomas Theil, CDBS
Primary cilia are small protrusions from the cell surface that act as cellular antennas to control cell signaling during development and in tissue homeostasis. Structural and functional defects in cilia result in a group of disorders known as ciliopathies and can affect every organ system. The image shows primary cilia emanating from neural stem cells in a human brain organoid. It is stained for INPP5e (green fluorophore) and gamma-Tubulin (red fluorophore) and counterstained with DAPI. The image was taken with the 60x objective at the Nikon confocal microscope in the HRB imaging facility.
14. Toxoplasma Bean
Dr Kseniia Bondarenko, University of Edinburgh
Two Toxoplasma gondii parasites sitting inside a human skin cell next to its big blue nuclei. The protein-rich parasitic and host cell parts are visible in red (594-NHS-ester, brightness corresponds to the protein content), parasite membrane in magenta (inner membrane complex, anti-GAP45), and nuclei in blue (DNA stain). Infected cells were gelled and expanded 4.5x using Ultrastructure Expansion Microscopy (U-ExM) and then imaged with Nikon Ti2 CSU-W1 Spinning Disk confocal microscope, 100x, at COIL imaging facility. Expansion Microscopy is a relatively new technique which allows increasing the spatial resolution through the physical expansion of the sample, bringing super-resolution quality to regular epifluorescence or confocal microscopes. U-ExM allows investigation of protein localization in Toxoplasma parasites with a spatial resolution of ~65 nm using a conventional confocal setup. We use this technique to clarify and discover new locations of proteins that are either upregulated or change their location when switching to the chronic state of toxoplasmosis. Model organism: human foreskin fibroblasts infected with ME49 strain of Toxoplasma gondii (tachyzoites). Prep: infected cells were fixed 24h post-infection using 4% formaldehyde and then stained against inner membrane complex using anti-GAP45, followed by incubation with a non-selective protein marker conjugated 594-NHS-ester, and Hoechst 33342 DNA stain.
15. Fusing PHOSPHO1 and MNeonGreen: Cellular or Solar Flares
Charlotte Clews, The Roslin Institute
PHOSPHO1 (a small phosphatase critical to the poorly understood process of bone mineralisation) was fused to MNeonGreen (488) in a transfection plasmid, and stably integrated into the genome of mouse osteoblast cell line MC3T3, in order to visualise the intracellular processing of the protein, and use super-resolution fluorescence imaging to observe PHOSPHO1 containing matrix vesicles and their release into the osteoblastic ECM, as has never been previously achieved. Spherical PHOSPHO1 objects appear to radiate from the perinuclear region (resembling, in this cell, solar flares…) and spread towards the periphery of the cell, as the culture prepares to mineralise the ECM. Fluorophores used include PHOSPHO1-MNeonGreen (488) with costains including Phalloidin-AlexaFluor594, Hoescht (405) and imaged using a Zeiss 880 with laser scanning confocal microscope in super resolution mode with airyscan processing. Scale bar = 20 µm
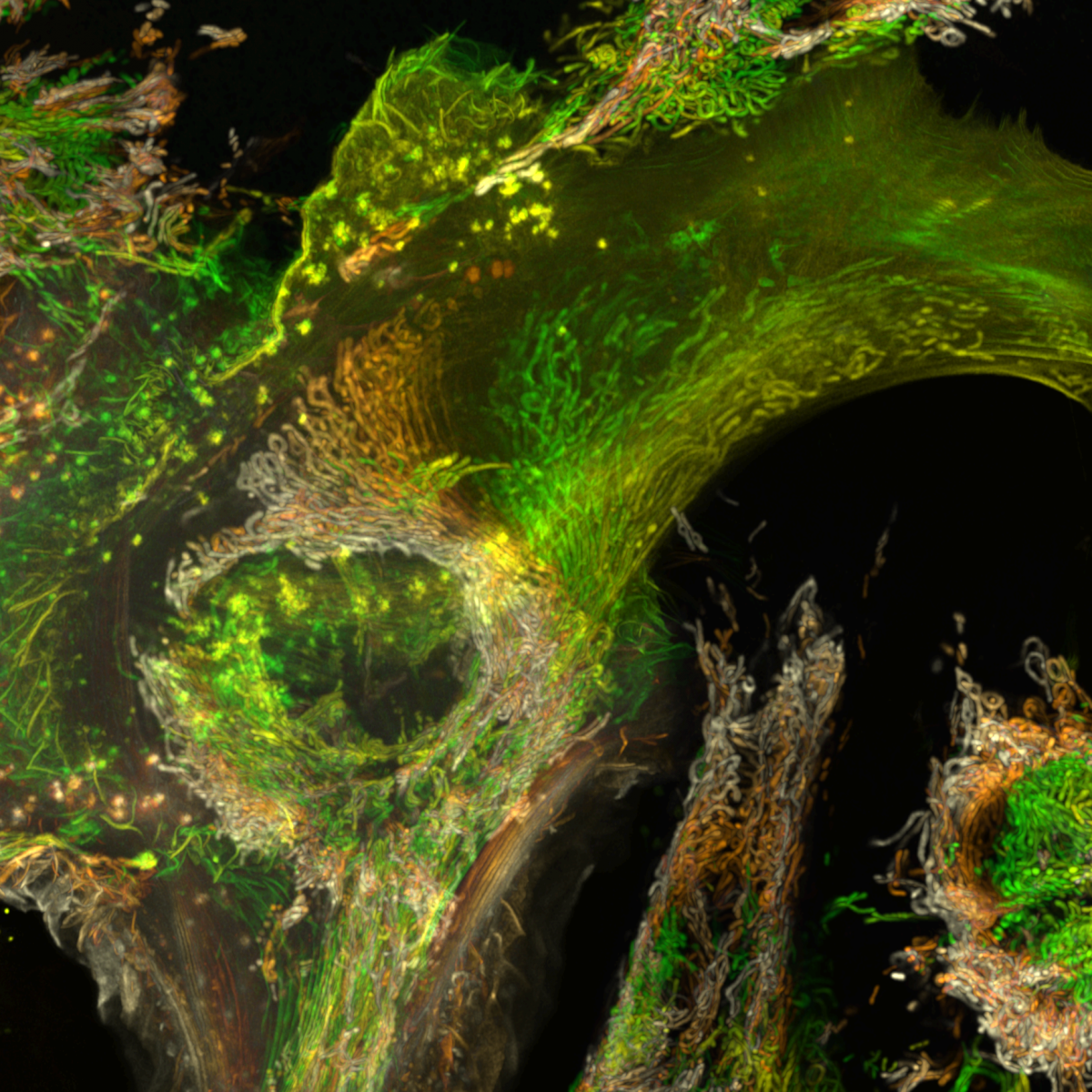
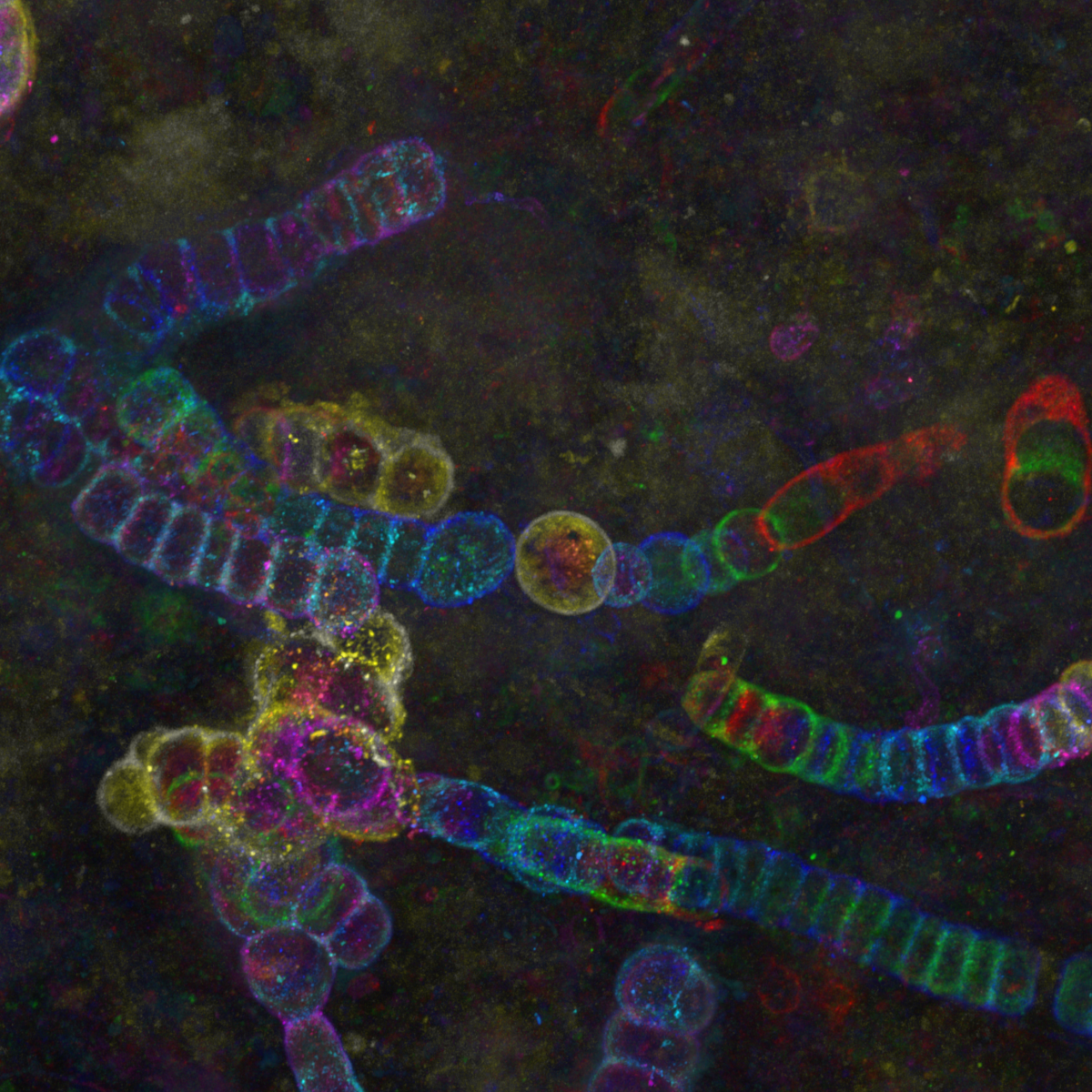
16. Shedding light on a 407 million year-old time capsule : Cyanobacteria from the Aberdeenshire
Dr Frédéric Fercoq, Muséum National d'Histoire Naturelle, Paris (France)
Color-coded maximum intensity projection image of cyanobacteria from the Lower Devonian Rhynie chert, Aberdeenshire (UK), observed by confocal microscopy. The rock thin section from the Palaeobotany Collection, Bibliothèque de Sorbonne Université, Paris (France) was observed at the imaging facility of the Muséum National d’Histoire Naturelle, Paris.
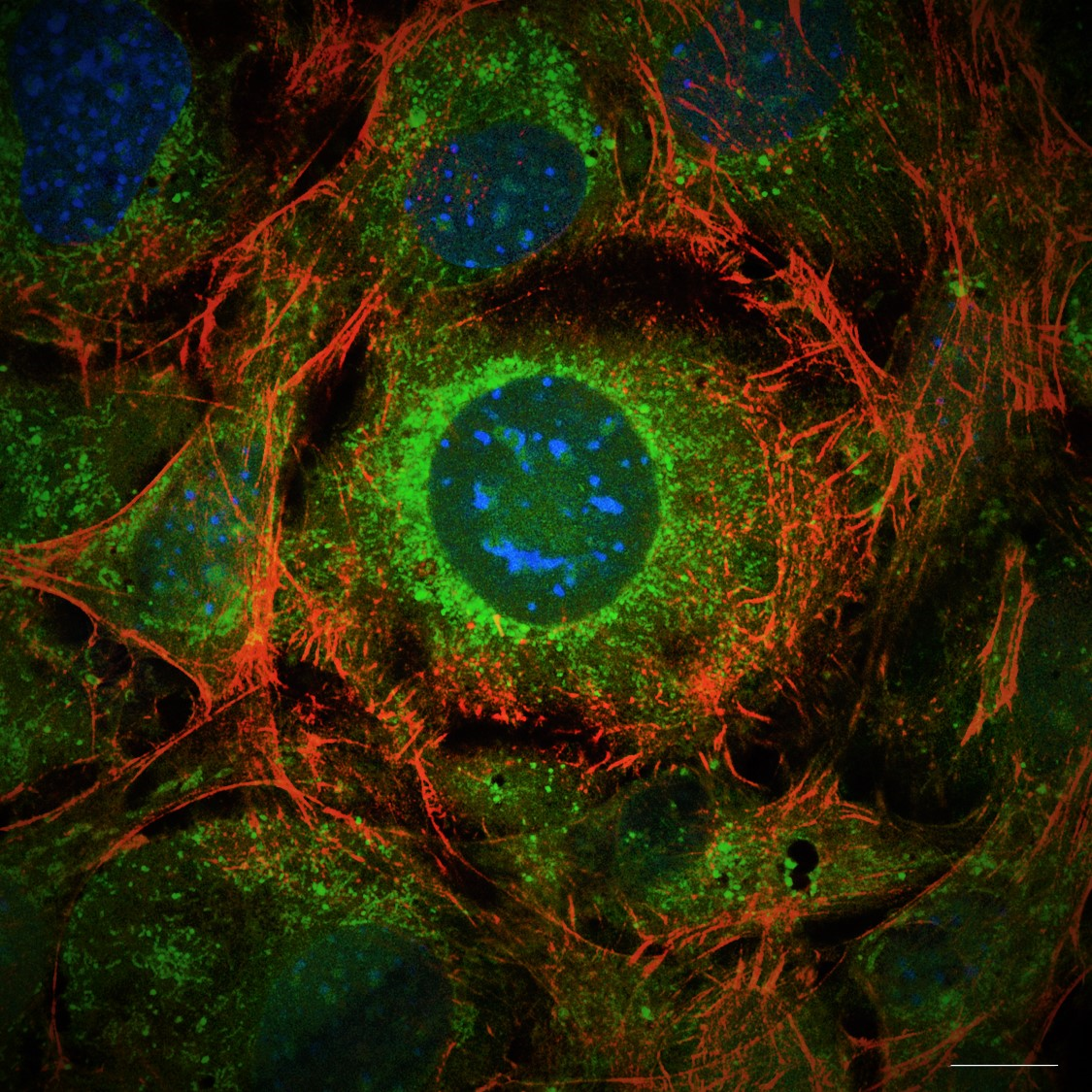
17. Fusing PHOSPHO1-MNeonGreen: vesicle release or new spring leaves?
Charlotte Clews, The Roslin Institute
PHOSPHO1 (a small phosphatase critical to the poorly understood process of bone mineralisation) was fused to MNeonGreen (488) in a transfection plasmid, and transiently transfected into mouse osteoblast cell line MC3T3, to visualise the process of calcium phosphate based mineralisation, as happens in bone. His process is thought to occur via small 50 – 200 nm sized extracellular vesicles (matrix vesicles), of which PHOSPHO1 is a marker, however these have not yet been observed via light microscopy in the published literature. This image shows PHOSPHO1 containing extracellular objects, within the correct size of matrix vesicles, and their release into the osteoblastic ECM via very long, filopodia-esque extensions, which appear to be rooted in the actin cytoskeleton. These extensions, dotted with PHOSPHO1-MNeonGreen positive puncta, grow out from the cell membrane (appearing like the branches/leaves of a tree in this image) into the ECM. Fluorophores used include PHOSPHO1-MNeonGreen (488) with Phalloidin-AlexaFluor594, and imaged using a Nikon Hybrid SORA / N-SIM system (The University of Edinburgh, Institute of Genetics and Cancer). Scale bar = 4 µm

Get in touch with the Scottish Microscopy Society!
Feel free to contact us or join our mailing list using the contact form below.